Welcome to uORFlight !
About uORFlight
Upstream open reading frames (uORFs) are prevalent in eukaryotic mRNAs. They act as a translational control element for precisely tuning the expression of the downstream major open reading frame (mORF). uORF variation has been clearly associated with several human diseases. In contrast, natural uORF variants in plants have not ever been identified or linked with any phenotypic changes. The paucity of such evidence encouraged us to generate this database-uORFlight (http://uorflight.whu.edu.cn). It facilitates the exploration of uORF variation among different splicing models of Arabidopsis and rice genes. Most importantly, users can evaluate uORF frequency among different accessions at the population scale and find out the causal single nucleotide polymorphism (SNP) or insertion/deletion (INDEL) which can be associated with phenotypic variation through database mining or simple experiments. Such information will help to make hypotheses of uORF function in plant development or adaption to changing environments on the basis of the cognate mORF function. This database also curates plant uORF relevant literature into distinct groups. To be broadly interesting, our database expands uORF annotation into more species of fungi (Botrytis cinerea, Saccharomyces cerevisiae), plant (Brassica napus, Glycine max, Gossypium raimondii, Medicago truncatula, Solanum lycopersicum, Solanum tuberosum,Triticum aestivum and Zea mays), metazoan (Caenorhabditis elegans and Drosophila melanogaster) and vertebrate (Homo sapiens, Mus musculus and Danio rerio). Therefore, uORFlight will light up the runway toward how uORF genetic variation determines phenotypic diversity and advance our understanding of translational control mechanisms in eukaryotes.
Database structure
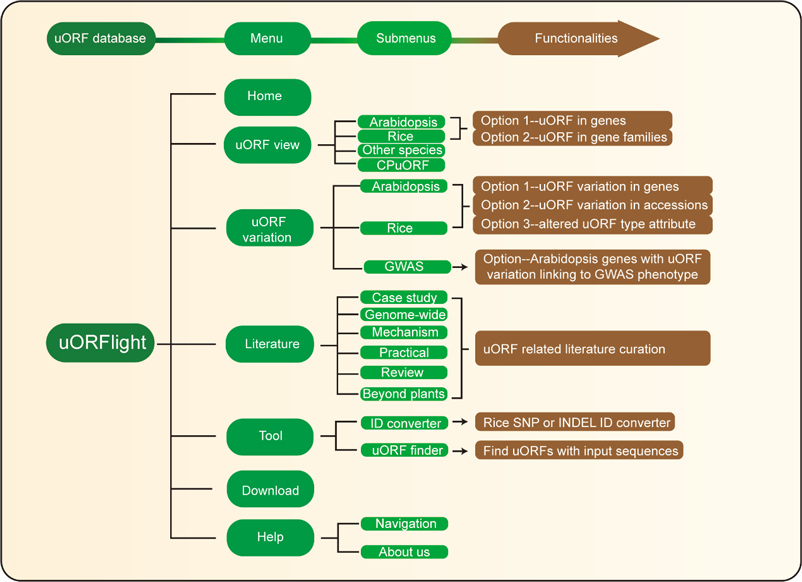
Home menu contains the background information of uORFlight database including organisms, and definition of uORF attributes and variants. uORF view menu has four submenus to browse and search uORF, including in the reference genomes of Arabidopsis Col-0, Rice Nipponbare and other species. CPuORF is also included in this menu. In Arabidopsis and Rice submenus, Option 1 and Option 2 are provided to individually and bulk retrieve uORF information, respectively. uORF variation menu is used to compare uORF variation among different splicing models in the reference genome (Option 1) or among the selected accessions (Option 2) and to bulk retrieve genes with altered uORF types (Option 3). Literature menu curates plant uORF relevant literature into distinct groups. Tool menu provides ID converter and uORF finder with the former to transform SNP and INDEL variation identity used in different external databases, and with the later to search ATG or Non-ATG initiated uORFs in a given cDNA sequence. Help menu contains Navigation submenu to explain the main conclusion on each result page.
About uORF
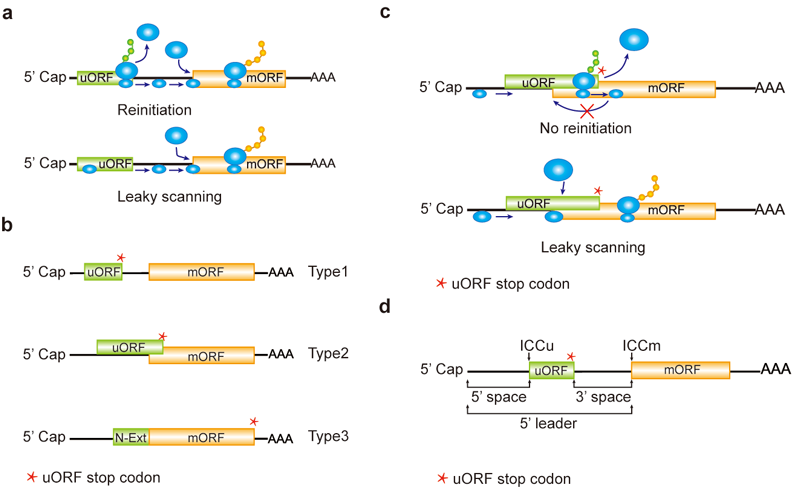
a, Reinitiation and leaky scanning models. In the reinitiation model, the 80S ribosomal subunit will separate after translating the uORF and the 40S subunit remains associated with mRNA, regaining fresh eIF2 ternary complex and other unknown reinitiation factors to translate the mORF. In the leaky scanning model, the uORF initiation codon is bypassed by the scanning complex, which will ignore the uORF and translate the mORF. b, uORF types. uORFs are divided into Types1-3 with respect to the position of uORF stop codon relative to the mORF. N-Ext, N extension. c, Type2 uORF-controlled mORF translation is only favored by leaky scanning. Overlap between the Type2 uORF and mORF makes reinitiation of the mORF impossible after translation of the uORF. d, uORF positional information on cDNA. The mORF is flanked by the 5’ leader and 3’ UTR (3’ untranslated region). 5’ and 3’ space are used to describe the distance from Cap to uORF AUG and from uORF stop codon to mORF AUG, respectively. The sequence from -3 to +4 relative to the AUG initiation codon (A as +1) corresponding to the Kozak consensus (A/GCCAUGG) position is termed as initiation codon context (ICC) with ICCu and ICCm for uORF and mORF, respectively.
Data source
1. Arabidopsis Col-0 accession (Ensemble V39; TAIR10) and rice Nipponbare cultivar (MSU V7) is used reference genomes for dicot and monocot uORF analysis, respectively.
2. VCF (variant call format) files of Arabidopsis 1135 accessions are from the 1001 Genomes Project and rice 3k varieties are from the 3000 Rice Genomes Project.
3. Other data are from Ensemble.
Citation
Niu R, Zhou Y, Zhang Y, et al. uORFlight: a vehicle toward uORF-mediated translational regulation mechanisms in eukaryotes. Database (Oxford). 2020;2020:baaa007. doi:10.1093/database/baaa007